📃 Pre-print • 👾 Discord bot • 🤗 Hugging Face
𝕏 @precious_gpt
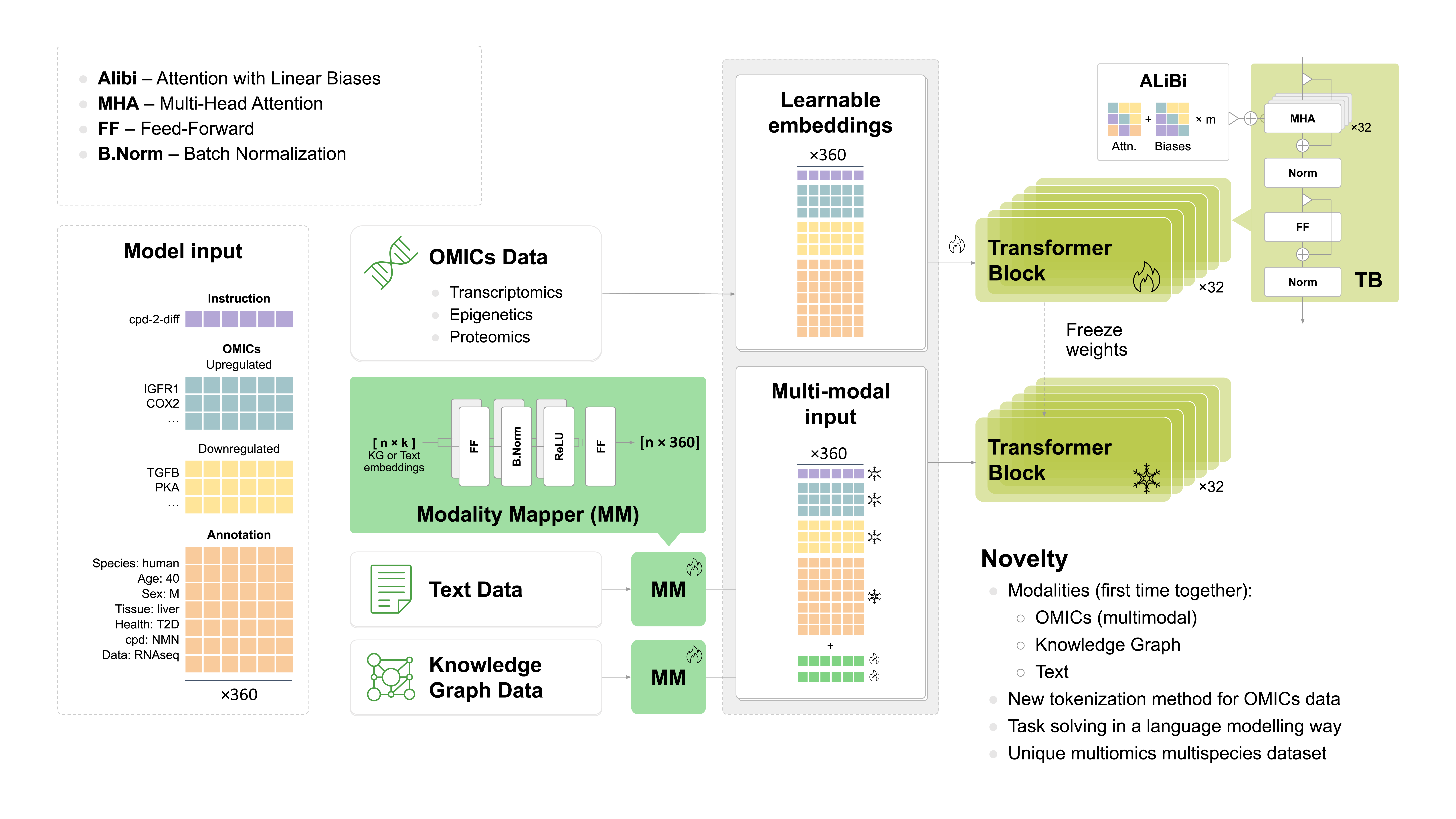
Python wrappers and utility classes for interacting with Precious3GPT (P3GPT), a multimodal transformer model for biomedical research and drug discovery.
P3GPT is a unique multimodal language model trained on:
- 1.2MM omics data points
- Knowledge graphs
- Biomedical texts (PubMed)
The model can simulate experiments using:
- 3 species
- 569 tissues and cell lines
- 635 health conditions
- 22k small molecules
The handlers in this repository enable easy:
- Digital case-control studies: Generate differential expression between conditions (young vs old, healthy vs diseased)
- Chemical screening simulation: Predict compound effects across tissues
- Multi-omics analysis: Get transcriptomic, epigenomic, or proteomic signatures for your experiment
- Python 3.11
- CUDA 12.4 with compatible NVIDIA drivers
- 16GB+ GPU memory recommended
git clone https://github.com/insilicomedicine/precious3-gpt.git
cd precious3-gpt
# Create conda environment
conda env create -f environment.yml
conda activate p3gptfrom handlers.p3_multimodal_handler import HandlerFactory
from handlers.screening import TopTokenScreening
# Initialize handler
handler = HandlerFactory.create_handler('endpoint', device='cuda:0')
# A handler enables a smooth and easy interaction with the P3GPT
# The handler will depend on the type of experiment you want to run
# Use the "endpoint" handler for aging- and disease-related studies
screen = TopTokenScreening(handler)
# Configure the screening grid
screen_grid = {
'tissue': ['whole blood', 'lung'],
'dataset_type': ['proteomics'],
'efo': ["", "EFO_0000768"],
'case': ["70.0-80.0", ""],
'control': ["19.95-25.0", ""]
}
# Add your screening grid
screen.add_grid(screen_grid)
# Generate 250 up-/down-regulated proteins for each grid point
screen(top_k=250)
# Save your screening DEGs as a TSV
screen.result_to_df.to_csv("./screening_output.txt", sep='\t', index = False)For a complete example analyzing aging signatures across multiple tissues and species, see this notebook.
P3GPT supports three execution modes:
meta2diff: Generate differentially expressed genes between conditions (currently the only tested and supported mode)diff2compound: Identify compounds that could induce given expression changesmeta2diff2compound: Combines both modes - generate expression profile and find matching compounds
For more details about P3GPT's capabilities and usage, please visit the original Hugging Face repository.
- Fork the repository
- Create your feature branch (
git checkout -b feature/amazing-feature) - Commit your changes (
git commit -m 'Add amazing feature') - Push to the branch (
git push origin feature/amazing-feature) - Open a Pull Request
This project is licensed under the MIT License - see the LICENSE file for details.
Cite the original preprint and model if you use the materials from this repo:
@article {Galkin2024.07.25.605062,
author = {Galkin, Fedor and Naumov, Vladimir and Pushkov, Stefan and Sidorenko, Denis and Urban, Anatoly and Zagirova, Diana and Alawi, Khadija M and Aliper, Alex and Gumerov, Ruslan and Kalashnikov, Aleksand and Mukba, Sabina and Pogorelskaya, Aleksandra and Ren, Feng and Shneyderman, Anastasia and Tang, Qiuqiong and Xiao, Deyong and Tyshkovskiy, Alexander and Ying, Kejun and Gladyshev, Vadim N. and Zhavoronkov, Alex},
title = {Precious3GPT: Multimodal Multi-Species Multi-Omics Multi-Tissue Transformer for Aging Research and Drug Discovery},
year = {2024},
doi = {10.1101/2024.07.25.605062},
publisher = {Cold Spring Harbor Laboratory},
journal = {bioRxiv}
}@misc {insilico_medicine_2024,
author = { {Insilico Medicine} },
title = { precious3-gpt-multi-modal (Revision 9e240ab) },
year = 2024,
url = { https://huggingface.co/insilicomedicine/precious3-gpt-multi-modal },
doi = { 10.57967/hf/2699 },
publisher = { Hugging Face }
}