-
Notifications
You must be signed in to change notification settings - Fork 28.5k
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
[SPARK-30699][ML][PYSPARK] GMM blockify input vectors #27473
Conversation
|
Test build #117976 has finished for PR 27473 at commit
|
|
Test build #122341 has finished for PR 27473 at commit
|
|
Test build #122342 has finished for PR 27473 at commit
|
|
Test build #122351 has finished for PR 27473 at commit
|
|
test on the first 1M rows in HIGGS: test code: import org.apache.spark.ml.clustering._
import org.apache.spark.storage.StorageLevel
import org.apache.spark.ml.linalg._
val df = spark.read.format("libsvm").load("/data1/Datasets/higgs/HIGGS.1m").repartition(1)
df.persist(StorageLevel.MEMORY_AND_DISK)
df.count
val gmm = new GaussianMixture().setSeed(0).setK(4).setMaxIter(2).setBlockSize(64)
gmm.fit(df)
val results = Seq(1, 4, 16, 64, 256, 1024, 4096).map { size => val start = System.currentTimeMillis; val model = gmm.setK(4).setMaxIter(20).setBlockSize(size).fit(df); val end = System.currentTimeMillis; (size, model, end - start) }
results.map(_._2.summary.numIter)
results.map(_._2.summary.logLikelihood)
results.map(_._3)Results WITHOUT native BLAS: It is surprising that there is a small performance regression on dense input: 105777 -> 106573 Results WITH native BLAS (OPENBLAS_NUM_THREADS=1): When OpenBLAS is used, it obtain about 3x speedup. Comparsion to Master (WITHOUT native BLAS): This PR keeps original behavior and performance if |
|
Test build #122358 has finished for PR 27473 at commit
|
|
@srowen I think GMM maybe a special case, I found than on dense input, it suffers a small regression. (Other impls like LoR/LiR will be accelerated significantly even without native-BLAS.) @xwu99 This is for GMM, I think it is similar to KMeans. I am happy if you can help reviewing this. |
|
Test build #122392 has finished for PR 27473 at commit
|
|
Test build #122398 has finished for PR 27473 at commit
|
|
retest this please |
|
Test build #122463 has finished for PR 27473 at commit
|
|
Test build #122466 has finished for PR 27473 at commit
|
|
Merged to master |
|
Let's don't merge without review or approval, @zhengruifeng |
|
Meta-comment - I have reviewed similar block-ification PRs and don't have a problem with them, and trust the judgment of @zhengruifeng as a result of the discussion. He now knows this code better than anyone. I know he's a committer and will "own" his work, including fixes if needed. I think this line of work is OK. |
|
This PR might be fine, and I understand ML side isn't reviewed pretty well either. But might be best to have somebody to follow his work up. If this becomes a pattern, I think it's a problem. |
|
@srowen Even if you reviewed some similar PRs from @zhengruifeng , could you explicitly give LGTM on each of the PRs before merge? Committers who saw PRs like #28473 would raise the same question as @HyukjinKwon . @zhengruifeng Please wait for LGTM before you merge a PR in the future. Committers should follow the review process strictly. |
|
@mengxr why not review them yourself? |
|
@mengxr OK, I will be more patient for reviewing. |
|
@zhengruifeng I trust that a committer would merge a PR if and only if he/she is confident with the change. However, a non-trivial change from the committer him/herself does not count. You have to find another committer to review and approve before merge, no matter how long you might wait. If no one reviews it, it means the community is not interested in the change. Your choice is to keep the PR open or close the PR, not merge. @srowen : @WeichenXu123 and I will take a look at the merged changes. |
| @@ -55,7 +55,7 @@ class MultivariateGaussian @Since("2.0.0") ( | |||
| */ | |||
| @transient private lazy val tuple = { | |||
| val (rootSigmaInv, u) = calculateCovarianceConstants | |||
| val rootSigmaInvMat = Matrices.fromBreeze(rootSigmaInv) | |||
| val rootSigmaInvMat = Matrices.fromBreeze(rootSigmaInv).toDense | |||
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
See comment in #29782
| @@ -81,6 +81,36 @@ class MultivariateGaussian @Since("2.0.0") ( | |||
| u - 0.5 * BLAS.dot(v, v) | |||
| } | |||
|
|
|||
| private[ml] def pdf(X: Matrix): DenseVector = { | |||
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
See comment in #29782
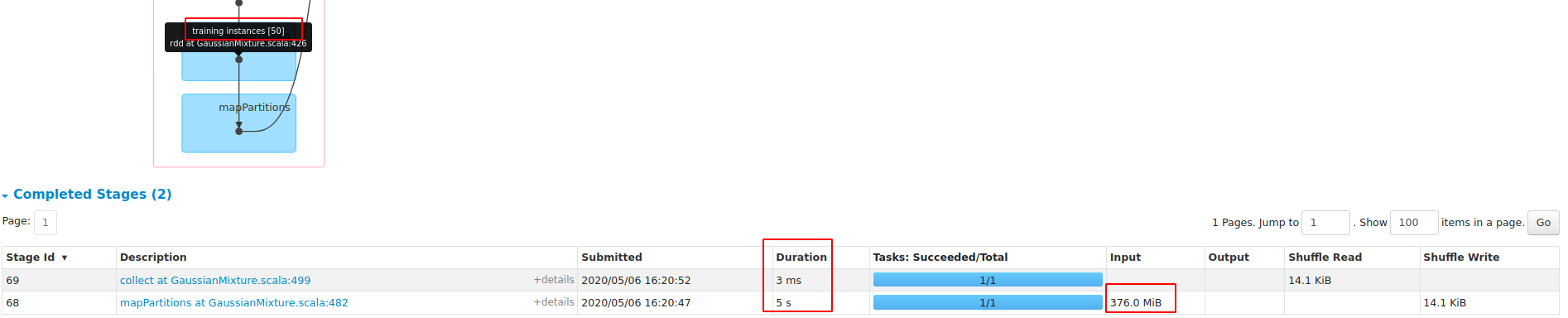
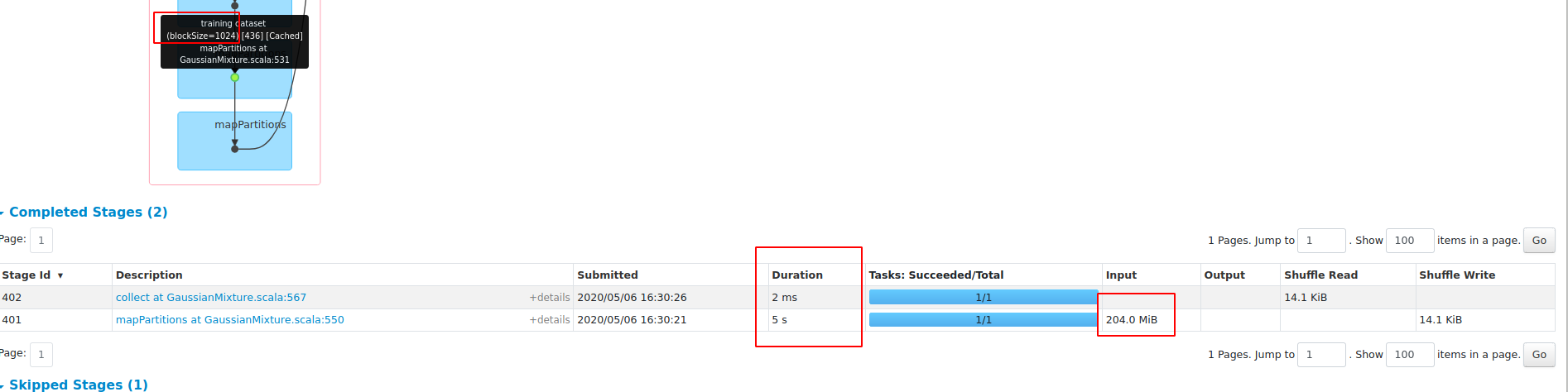
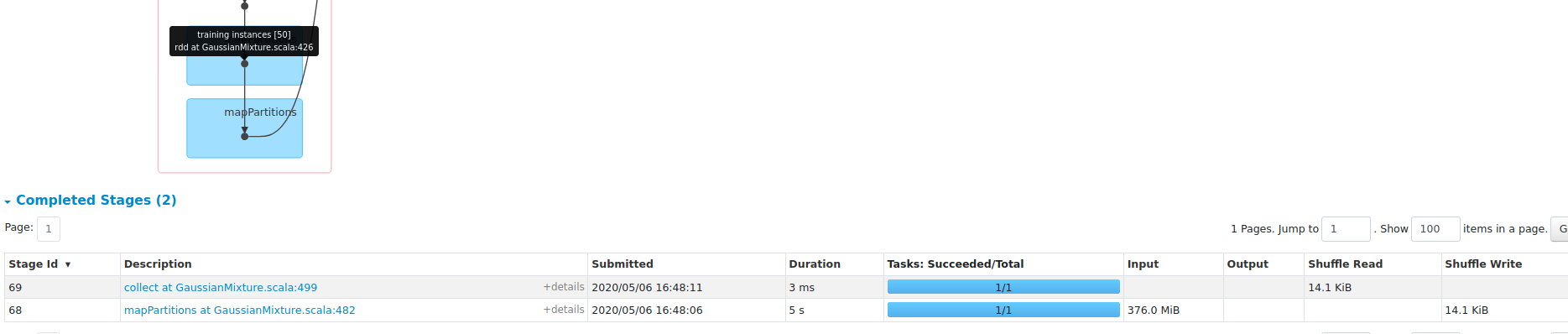
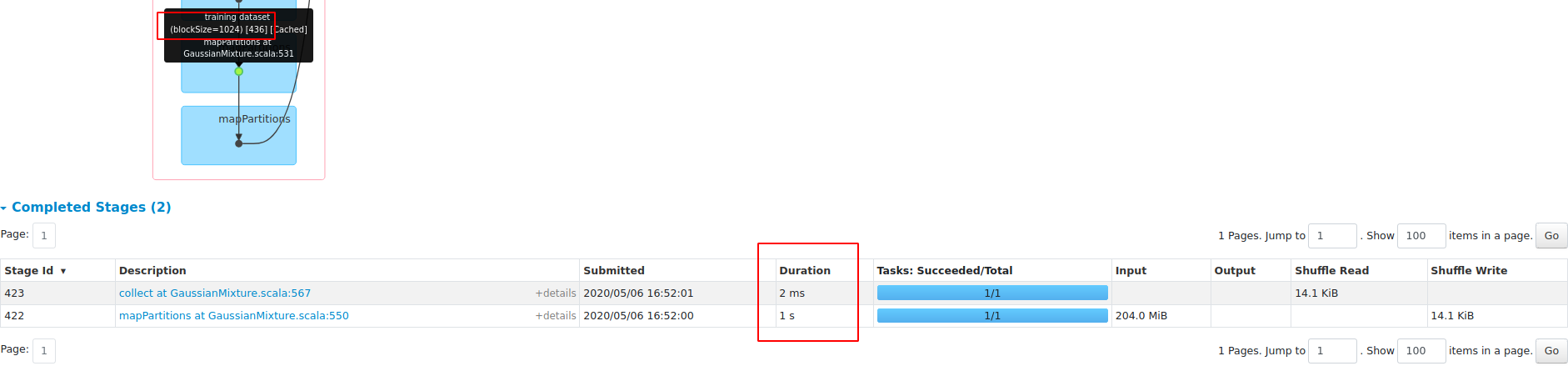
What changes were proposed in this pull request?
1, add new param blockSize;
2, if blockSize==1, keep original behavior, code path trainOnRows;
3, if blockSize>1, standardize and stack input vectors to blocks (like ALS/MLP), code path trainOnBlocks
Why are the changes needed?
performance gain on dense dataset HIGGS:
1, save about 45% RAM;
2, 3X faster with openBLAS
Does this PR introduce any user-facing change?
add a new expert param
blockSizeHow was this patch tested?
added testsuites