-
Notifications
You must be signed in to change notification settings - Fork 28.5k
Commit
This commit does not belong to any branch on this repository, and may belong to a fork outside of the repository.
[SPARK-3181][ML] Implement huber loss for LinearRegression.
## What changes were proposed in this pull request? MLlib ```LinearRegression``` supports _huber_ loss addition to _leastSquares_ loss. The huber loss objective function is: 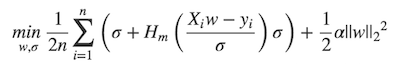 Refer Eq.(6) and Eq.(8) in [A robust hybrid of lasso and ridge regression](http://statweb.stanford.edu/~owen/reports/hhu.pdf). This objective is jointly convex as a function of (w, σ) ∈ R × (0,∞), we can use L-BFGS-B to solve it. The current implementation is a straight forward porting for Python scikit-learn [```HuberRegressor```](http://scikit-learn.org/stable/modules/generated/sklearn.linear_model.HuberRegressor.html). There are some differences: * We use mean loss (```lossSum/weightSum```), but sklearn uses total loss (```lossSum```). * We multiply the loss function and L2 regularization by 1/2. It does not affect the result if we multiply the whole formula by a factor, we just keep consistent with _leastSquares_ loss. So if fitting w/o regularization, MLlib and sklearn produce the same output. If fitting w/ regularization, MLlib should set ```regParam``` divide by the number of instances to match the output of sklearn. ## How was this patch tested? Unit tests. Author: Yanbo Liang <ybliang8@gmail.com> Closes #19020 from yanboliang/spark-3181.
- Loading branch information
1 parent
2a29a60
commit 1e44dd0
Showing
7 changed files
with
823 additions
and
65 deletions.
There are no files selected for viewing
150 changes: 150 additions & 0 deletions
150
mllib/src/main/scala/org/apache/spark/ml/optim/aggregator/HuberAggregator.scala
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -0,0 +1,150 @@ | ||
| /* | ||
| * Licensed to the Apache Software Foundation (ASF) under one or more | ||
| * contributor license agreements. See the NOTICE file distributed with | ||
| * this work for additional information regarding copyright ownership. | ||
| * The ASF licenses this file to You under the Apache License, Version 2.0 | ||
| * (the "License"); you may not use this file except in compliance with | ||
| * the License. You may obtain a copy of the License at | ||
| * | ||
| * http://www.apache.org/licenses/LICENSE-2.0 | ||
| * | ||
| * Unless required by applicable law or agreed to in writing, software | ||
| * distributed under the License is distributed on an "AS IS" BASIS, | ||
| * WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied. | ||
| * See the License for the specific language governing permissions and | ||
| * limitations under the License. | ||
| */ | ||
| package org.apache.spark.ml.optim.aggregator | ||
|
|
||
| import org.apache.spark.broadcast.Broadcast | ||
| import org.apache.spark.ml.feature.Instance | ||
| import org.apache.spark.ml.linalg.Vector | ||
|
|
||
| /** | ||
| * HuberAggregator computes the gradient and loss for a huber loss function, | ||
| * as used in robust regression for samples in sparse or dense vector in an online fashion. | ||
| * | ||
| * The huber loss function based on: | ||
| * <a href="http://statweb.stanford.edu/~owen/reports/hhu.pdf">Art B. Owen (2006), | ||
| * A robust hybrid of lasso and ridge regression</a>. | ||
| * | ||
| * Two HuberAggregator can be merged together to have a summary of loss and gradient of | ||
| * the corresponding joint dataset. | ||
| * | ||
| * The huber loss function is given by | ||
| * | ||
| * <blockquote> | ||
| * $$ | ||
| * \begin{align} | ||
| * \min_{w, \sigma}\frac{1}{2n}{\sum_{i=1}^n\left(\sigma + | ||
| * H_m\left(\frac{X_{i}w - y_{i}}{\sigma}\right)\sigma\right) + \frac{1}{2}\lambda {||w||_2}^2} | ||
| * \end{align} | ||
| * $$ | ||
| * </blockquote> | ||
| * | ||
| * where | ||
| * | ||
| * <blockquote> | ||
| * $$ | ||
| * \begin{align} | ||
| * H_m(z) = \begin{cases} | ||
| * z^2, & \text {if } |z| < \epsilon, \\ | ||
| * 2\epsilon|z| - \epsilon^2, & \text{otherwise} | ||
| * \end{cases} | ||
| * \end{align} | ||
| * $$ | ||
| * </blockquote> | ||
| * | ||
| * It is advised to set the parameter $\epsilon$ to 1.35 to achieve 95% statistical efficiency | ||
| * for normally distributed data. Please refer to chapter 2 of | ||
| * <a href="http://statweb.stanford.edu/~owen/reports/hhu.pdf"> | ||
| * A robust hybrid of lasso and ridge regression</a> for more detail. | ||
| * | ||
| * @param fitIntercept Whether to fit an intercept term. | ||
| * @param epsilon The shape parameter to control the amount of robustness. | ||
| * @param bcFeaturesStd The broadcast standard deviation values of the features. | ||
| * @param bcParameters including three parts: the regression coefficients corresponding | ||
| * to the features, the intercept (if fitIntercept is ture) | ||
| * and the scale parameter (sigma). | ||
| */ | ||
| private[ml] class HuberAggregator( | ||
| fitIntercept: Boolean, | ||
| epsilon: Double, | ||
| bcFeaturesStd: Broadcast[Array[Double]])(bcParameters: Broadcast[Vector]) | ||
| extends DifferentiableLossAggregator[Instance, HuberAggregator] { | ||
|
|
||
| protected override val dim: Int = bcParameters.value.size | ||
| private val numFeatures: Int = if (fitIntercept) dim - 2 else dim - 1 | ||
| private val sigma: Double = bcParameters.value(dim - 1) | ||
| private val intercept: Double = if (fitIntercept) { | ||
| bcParameters.value(dim - 2) | ||
| } else { | ||
| 0.0 | ||
| } | ||
|
|
||
| /** | ||
| * Add a new training instance to this HuberAggregator, and update the loss and gradient | ||
| * of the objective function. | ||
| * | ||
| * @param instance The instance of data point to be added. | ||
| * @return This HuberAggregator object. | ||
| */ | ||
| def add(instance: Instance): HuberAggregator = { | ||
| instance match { case Instance(label, weight, features) => | ||
| require(numFeatures == features.size, s"Dimensions mismatch when adding new sample." + | ||
| s" Expecting $numFeatures but got ${features.size}.") | ||
| require(weight >= 0.0, s"instance weight, $weight has to be >= 0.0") | ||
|
|
||
| if (weight == 0.0) return this | ||
| val localFeaturesStd = bcFeaturesStd.value | ||
| val localCoefficients = bcParameters.value.toArray.slice(0, numFeatures) | ||
| val localGradientSumArray = gradientSumArray | ||
|
|
||
| val margin = { | ||
| var sum = 0.0 | ||
| features.foreachActive { (index, value) => | ||
| if (localFeaturesStd(index) != 0.0 && value != 0.0) { | ||
| sum += localCoefficients(index) * (value / localFeaturesStd(index)) | ||
| } | ||
| } | ||
| if (fitIntercept) sum += intercept | ||
| sum | ||
| } | ||
| val linearLoss = label - margin | ||
|
|
||
| if (math.abs(linearLoss) <= sigma * epsilon) { | ||
| lossSum += 0.5 * weight * (sigma + math.pow(linearLoss, 2.0) / sigma) | ||
| val linearLossDivSigma = linearLoss / sigma | ||
|
|
||
| features.foreachActive { (index, value) => | ||
| if (localFeaturesStd(index) != 0.0 && value != 0.0) { | ||
| localGradientSumArray(index) += | ||
| -1.0 * weight * linearLossDivSigma * (value / localFeaturesStd(index)) | ||
| } | ||
| } | ||
| if (fitIntercept) { | ||
| localGradientSumArray(dim - 2) += -1.0 * weight * linearLossDivSigma | ||
| } | ||
| localGradientSumArray(dim - 1) += 0.5 * weight * (1.0 - math.pow(linearLossDivSigma, 2.0)) | ||
| } else { | ||
| val sign = if (linearLoss >= 0) -1.0 else 1.0 | ||
| lossSum += 0.5 * weight * | ||
| (sigma + 2.0 * epsilon * math.abs(linearLoss) - sigma * epsilon * epsilon) | ||
|
|
||
| features.foreachActive { (index, value) => | ||
| if (localFeaturesStd(index) != 0.0 && value != 0.0) { | ||
| localGradientSumArray(index) += | ||
| weight * sign * epsilon * (value / localFeaturesStd(index)) | ||
| } | ||
| } | ||
| if (fitIntercept) { | ||
| localGradientSumArray(dim - 2) += weight * sign * epsilon | ||
| } | ||
| localGradientSumArray(dim - 1) += 0.5 * weight * (1.0 - epsilon * epsilon) | ||
| } | ||
|
|
||
| weightSum += weight | ||
| this | ||
| } | ||
| } | ||
| } |
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
Oops, something went wrong.